| Feature | Summary |
|---|---|
| Tier_1.Exclusion_before_analysis | Not_excluded |
| Tier_2.Recover_all_40_SCGs | Yes |
| Tier_3.Adult_stool_from_well_sampled_countries | Yes |
| Tier_4.Adult_stool_from_healthy_subjects_not_currently_on_antibiotic | No, Yes |
| BodySite | Stool |
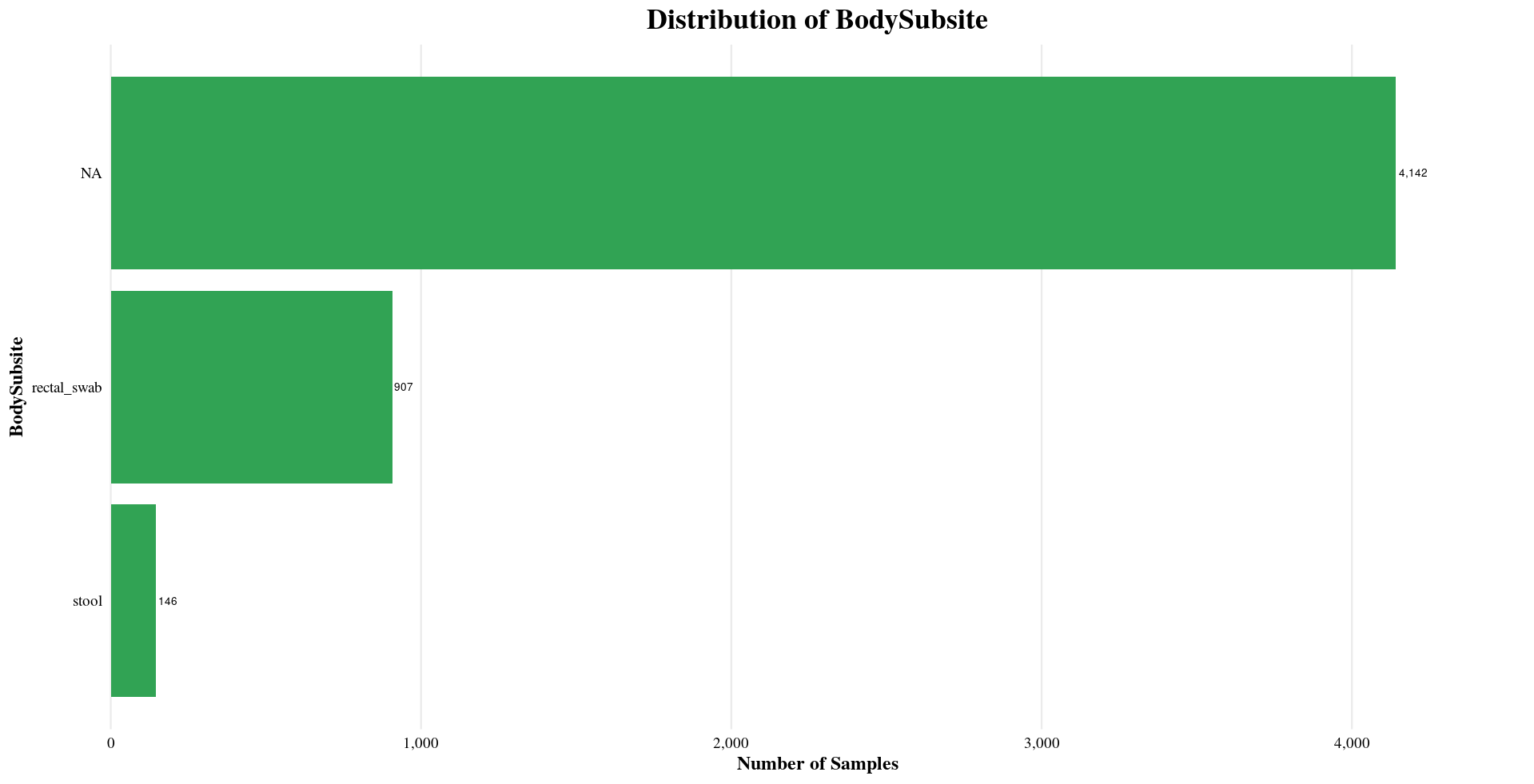
| BodySubsite | NA, stool, rectal_swab |
| AgeCategory | Adult, School age, Senior |
| Westernized | Yes, No |
| Country | ITA, SWE, FJI, MDG, CHN, DEU, KAZ, BGD, AUT, USA, CAN, FRA, DNK, ESP, NLD, MNG, PER, TZA, GBR, ISR |
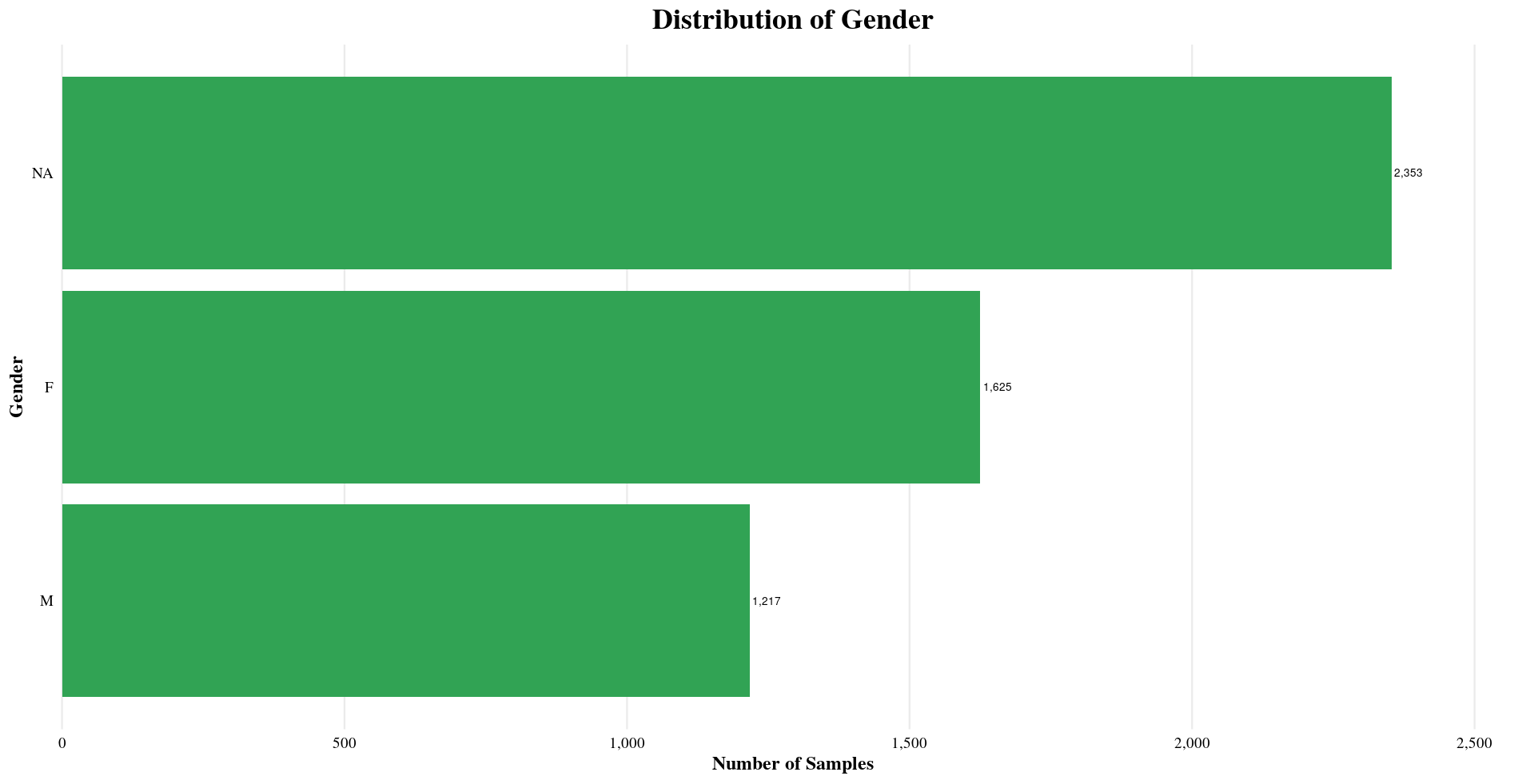
| Gender | F, NA, M |
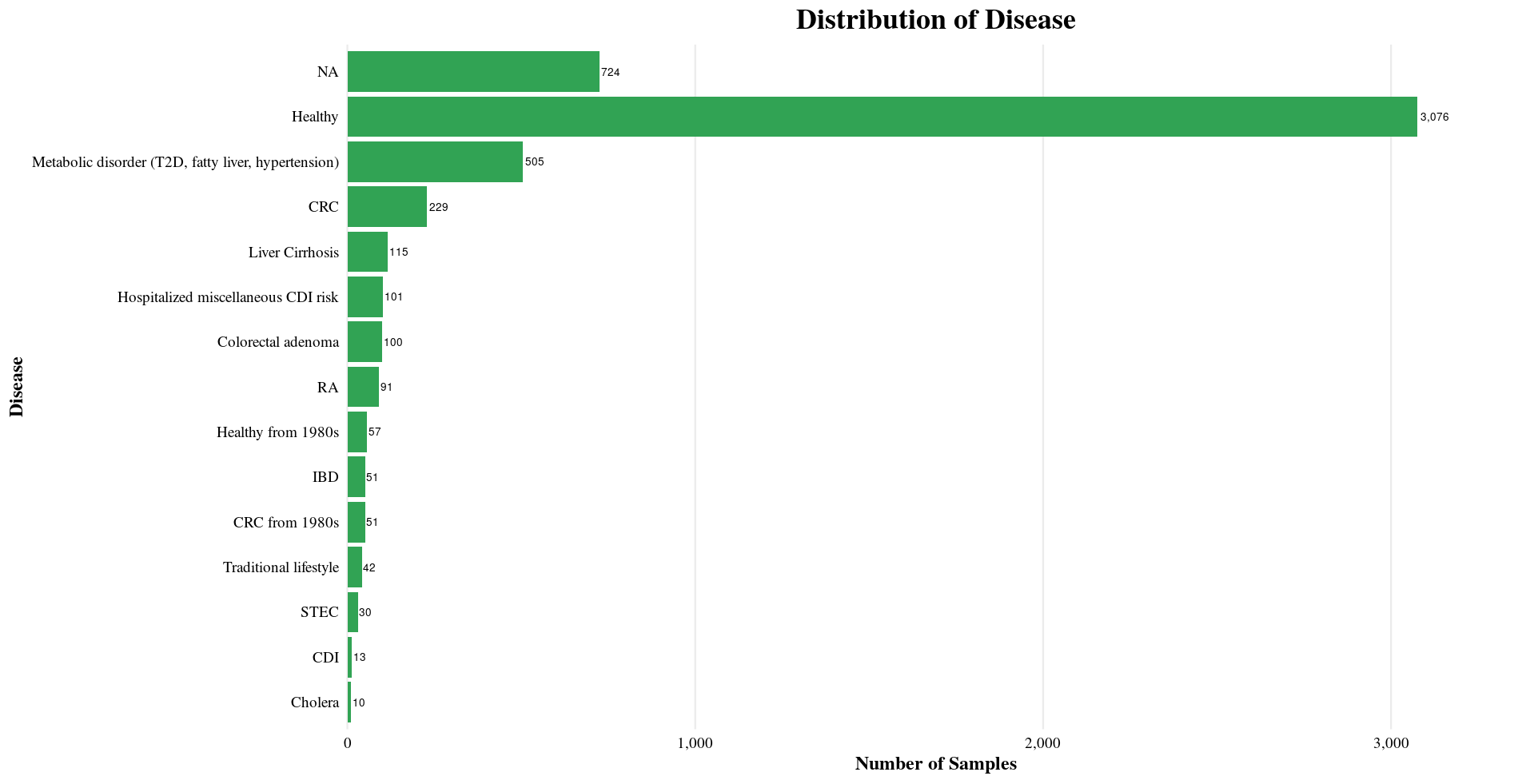
| Disease | Healthy, NA, RA, Cholera, CRC, Metabolic disorder, Colorectal adenoma, IBD, etc. |
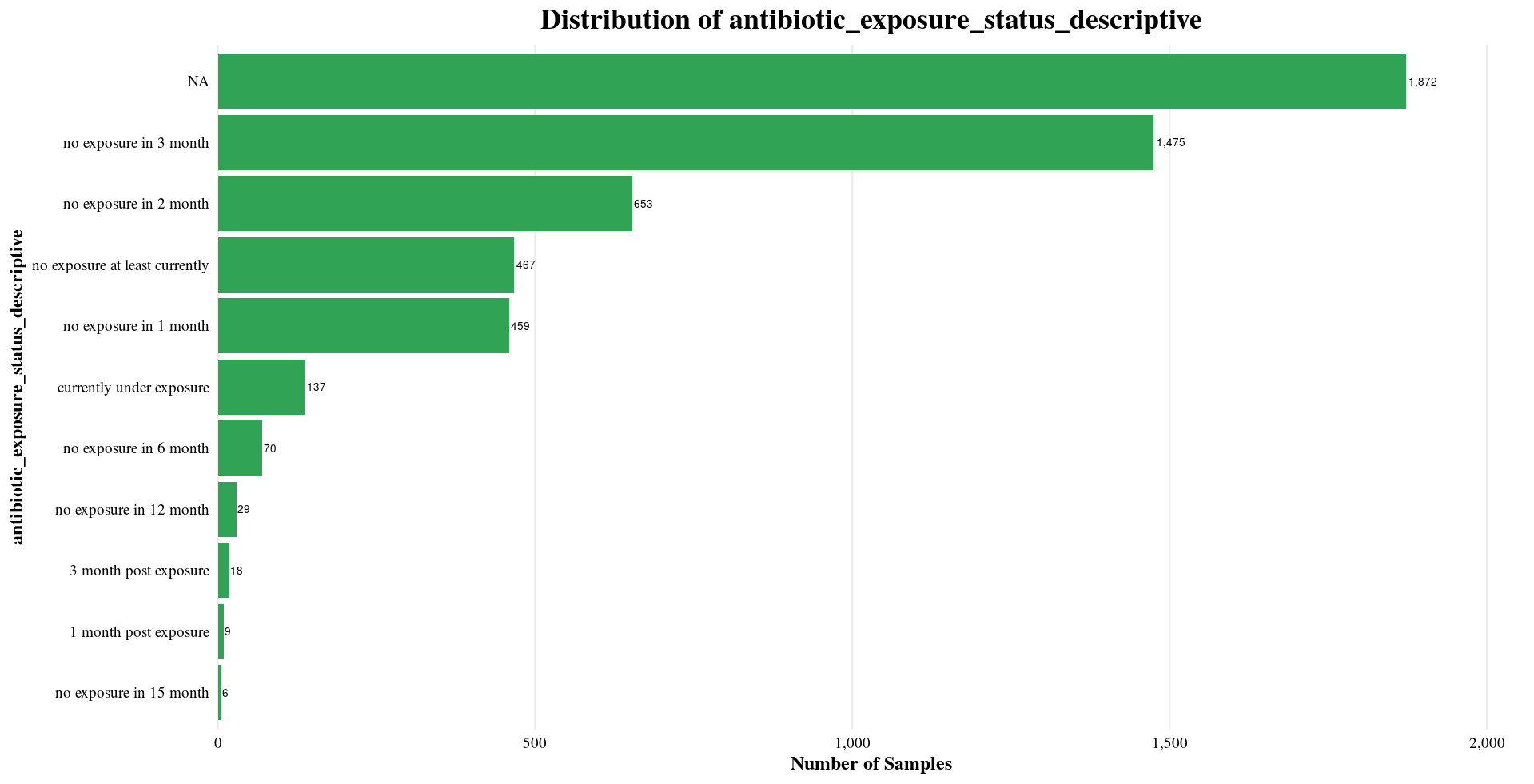
| antibiotic_exposure_status_descriptive | NA, no exposure in 6 months, no exposure at least currently, etc. |
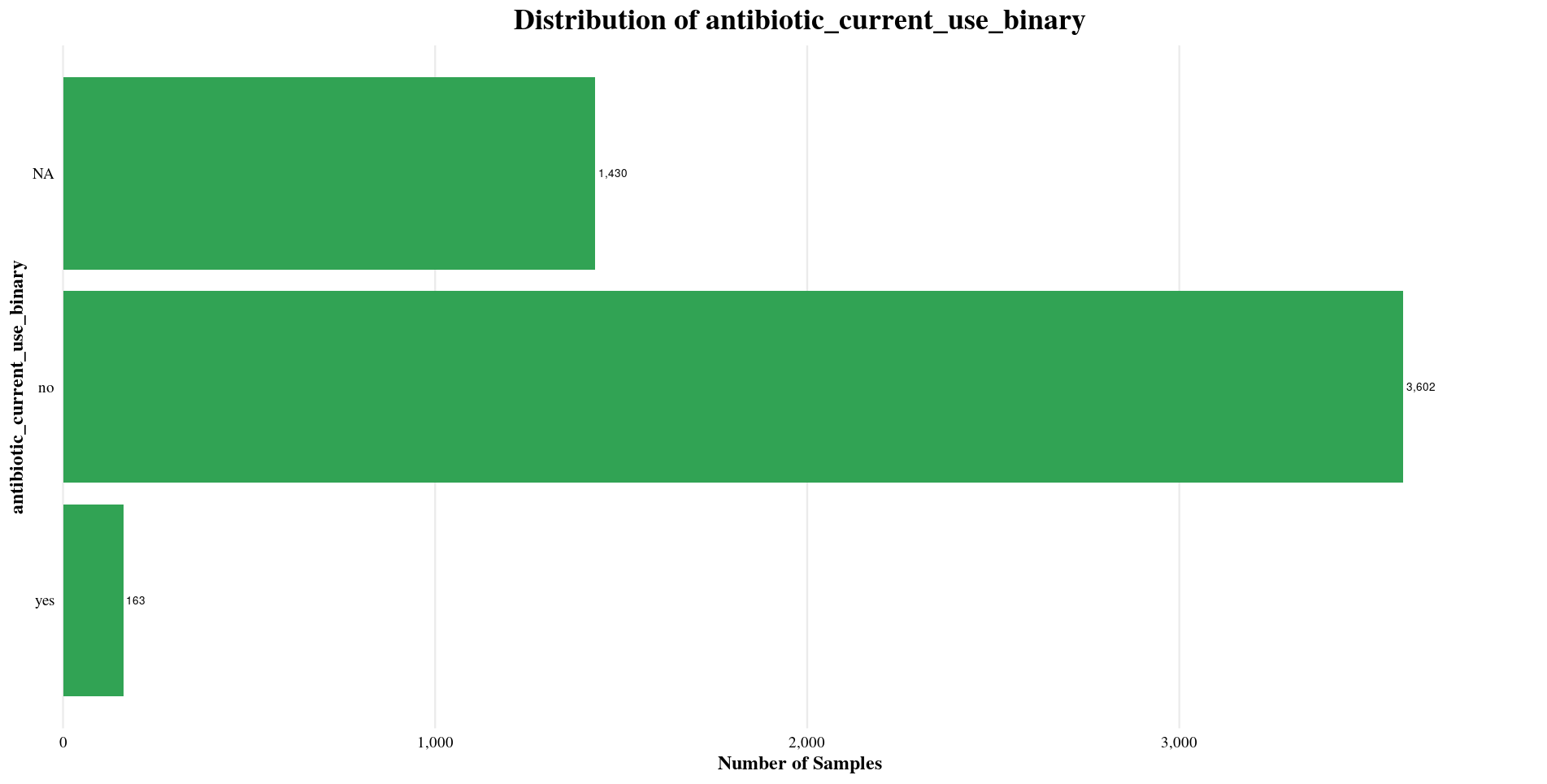
| antibiotic_current_use_binary | NA, no, yes |
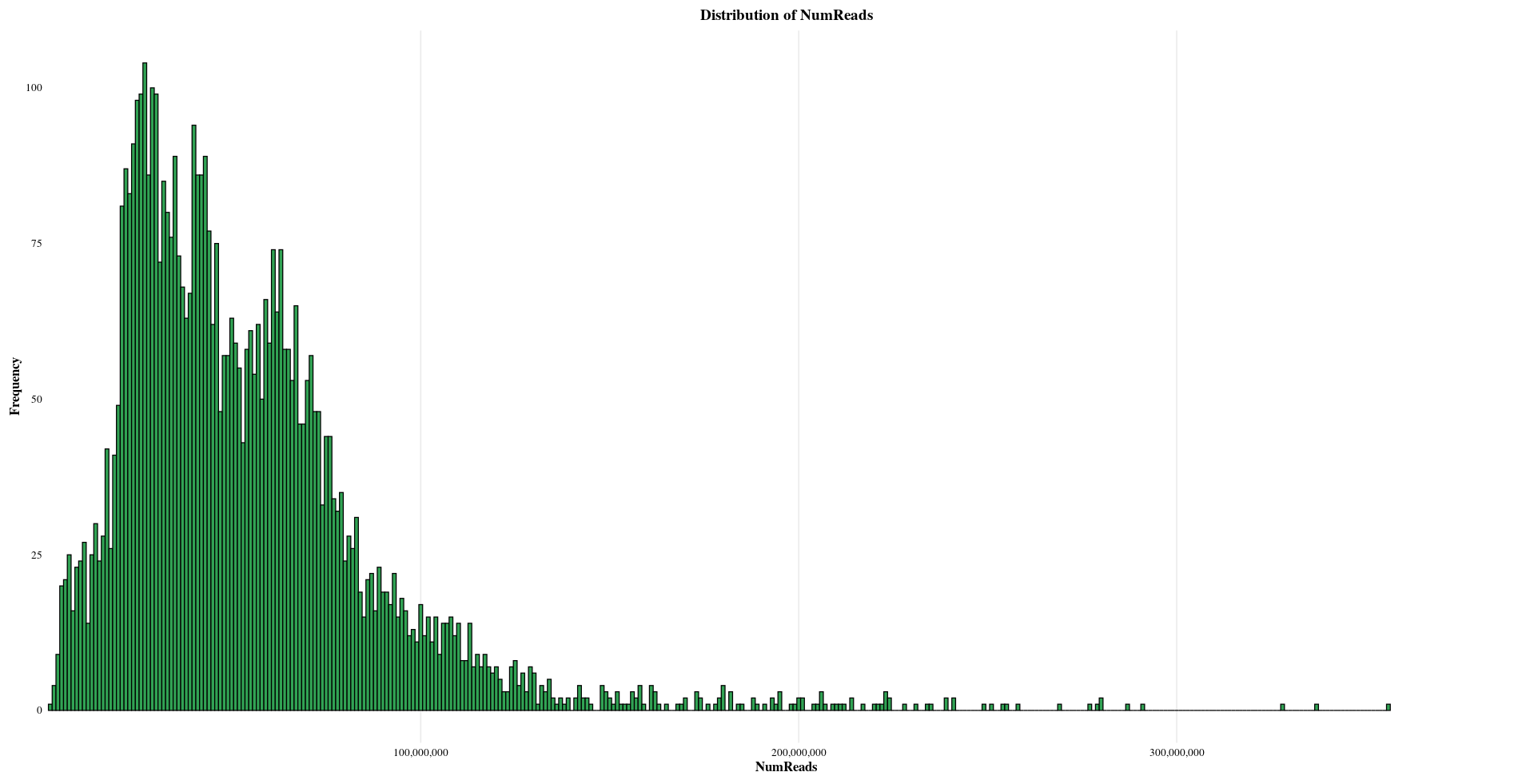
| NumReads | Mean: 53841103, Median: 46307042, Min: 2294514, Max: 356009214 |
| NumBases | Mean: 5268656695, Median: 4729928293, Min: 285954703, Max: 41957745000 |
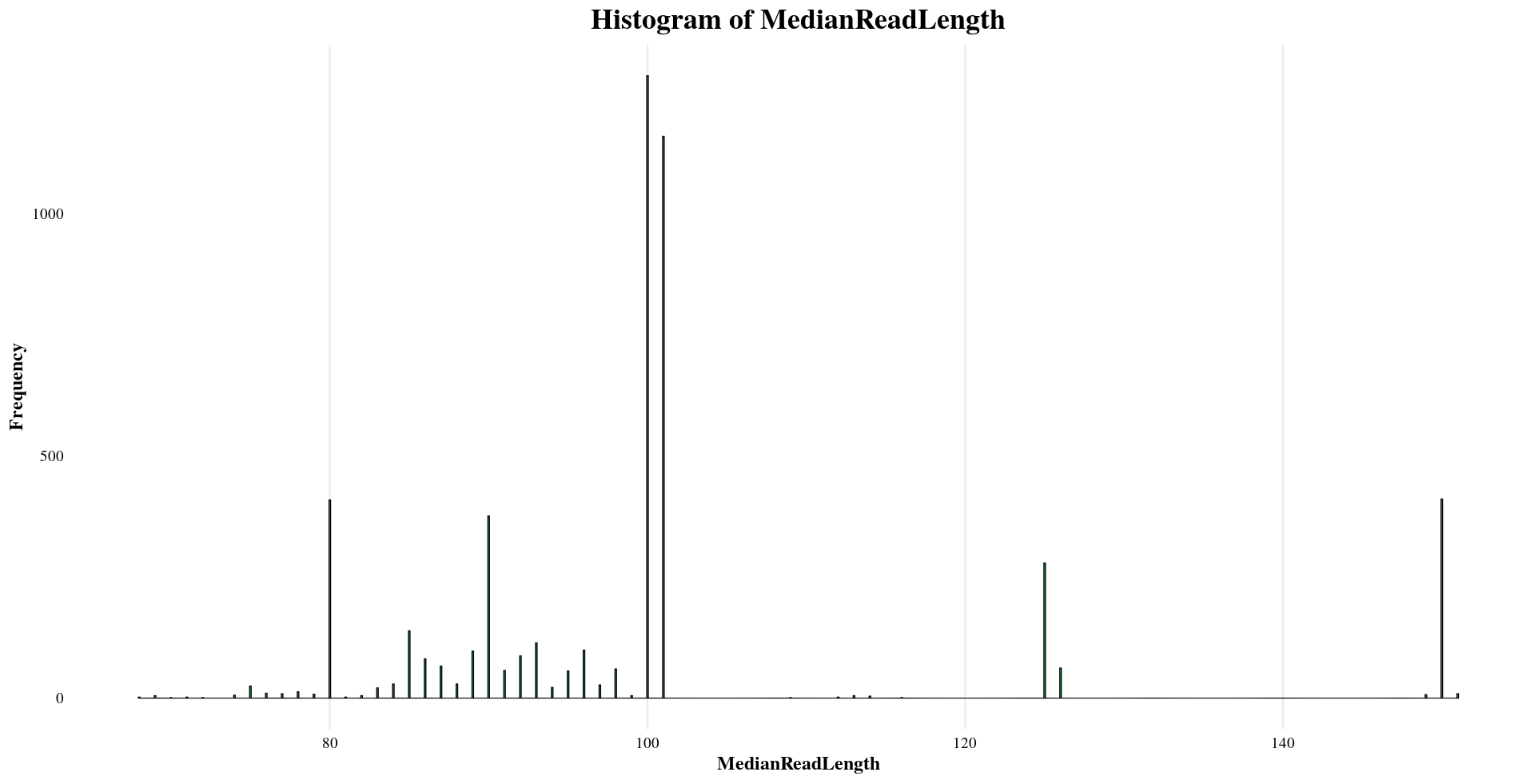
| MedianReadLength | Mean: 102, Median: 100, Min: 68, Max: 151 |
| AgeYears | Mean: 46, Median: 45, Min: 12, Max: 91 |
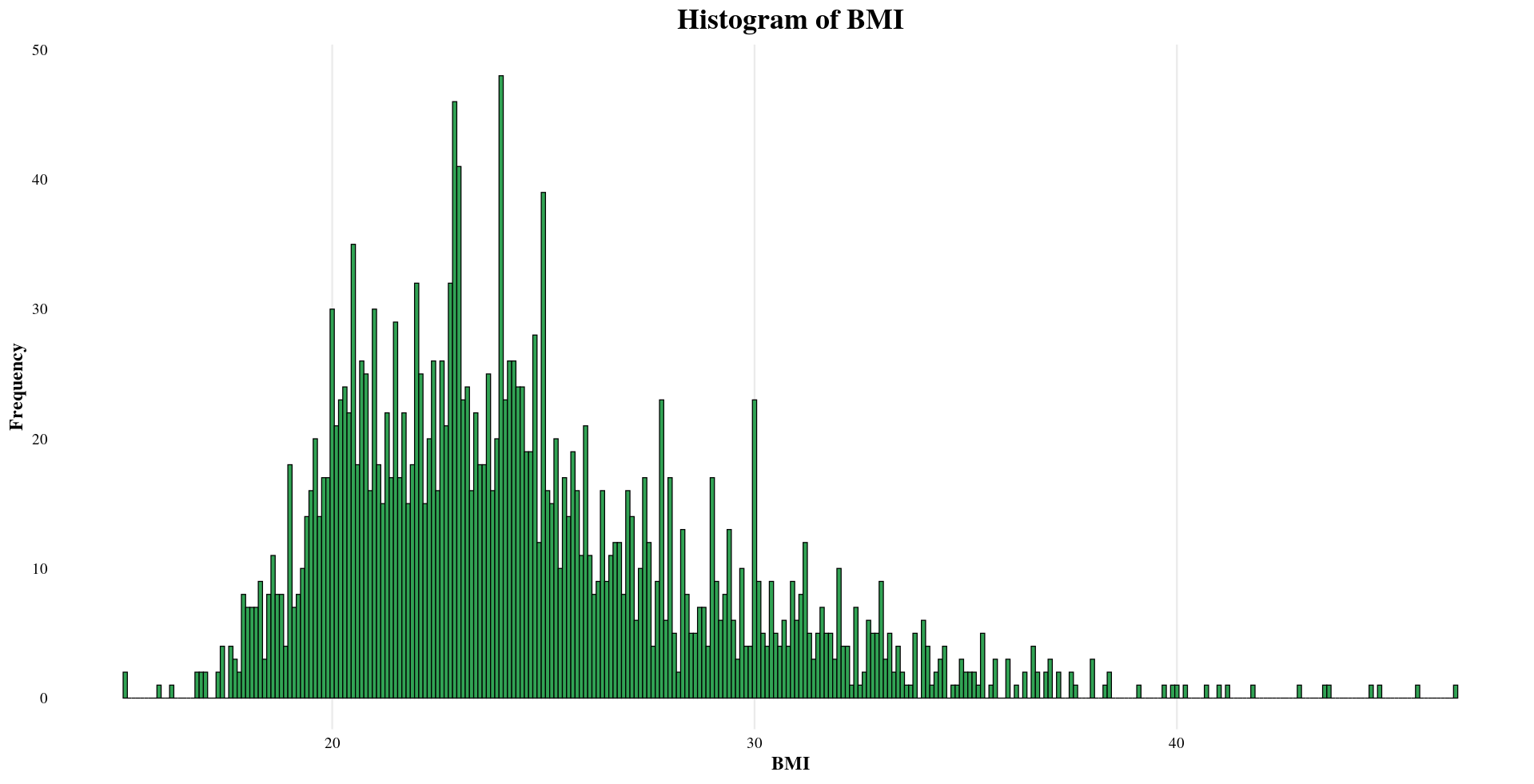
| BMI | Mean: 25, Median: 24, Min: 15, Max: 47 |
| InfantAge | Mean: 1, Median: 1, Min: 1, Max: 1 |
Data Summary
Data Source
This large-scale study includes a total of 9,251 samples collected from various human body sites and countries.
Aims of the study
This study explores how antibiotic use affects the abundance and diversity of resistance genes (ARGs) in the human gut.
Analyzing samples from healthy individuals, it finds links between national antibiotic usage and ARG prevalence, while also examining ARG transfer and resistotype patterns in the gut.
Methods: Data Collection & Compilation
Raw Illumina sequencing reads from NCBI or EMBL using Kingfisher-download
Metagenomic assemblies retrieved from the paper “Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle”
6104 adult gut metagenome samples from 20 countries after refinning
curatedMetagenomeData R package for retrieving the sample metadata
- Manually curated ARG families from CARD database (n=752)
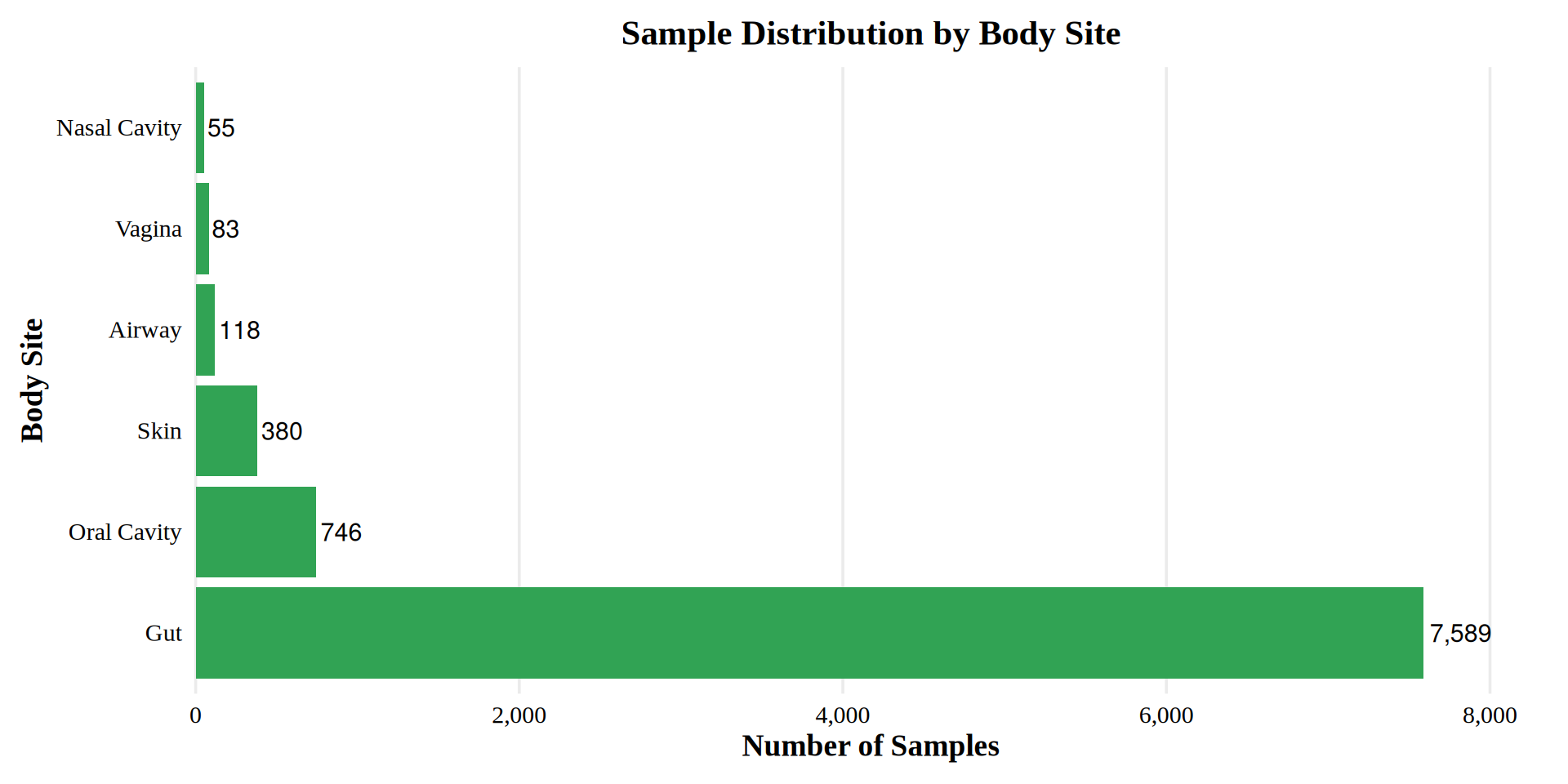
Sample Distribution by Body Site
The dataset comprises samples from six major body sites, with the distribution as follows:
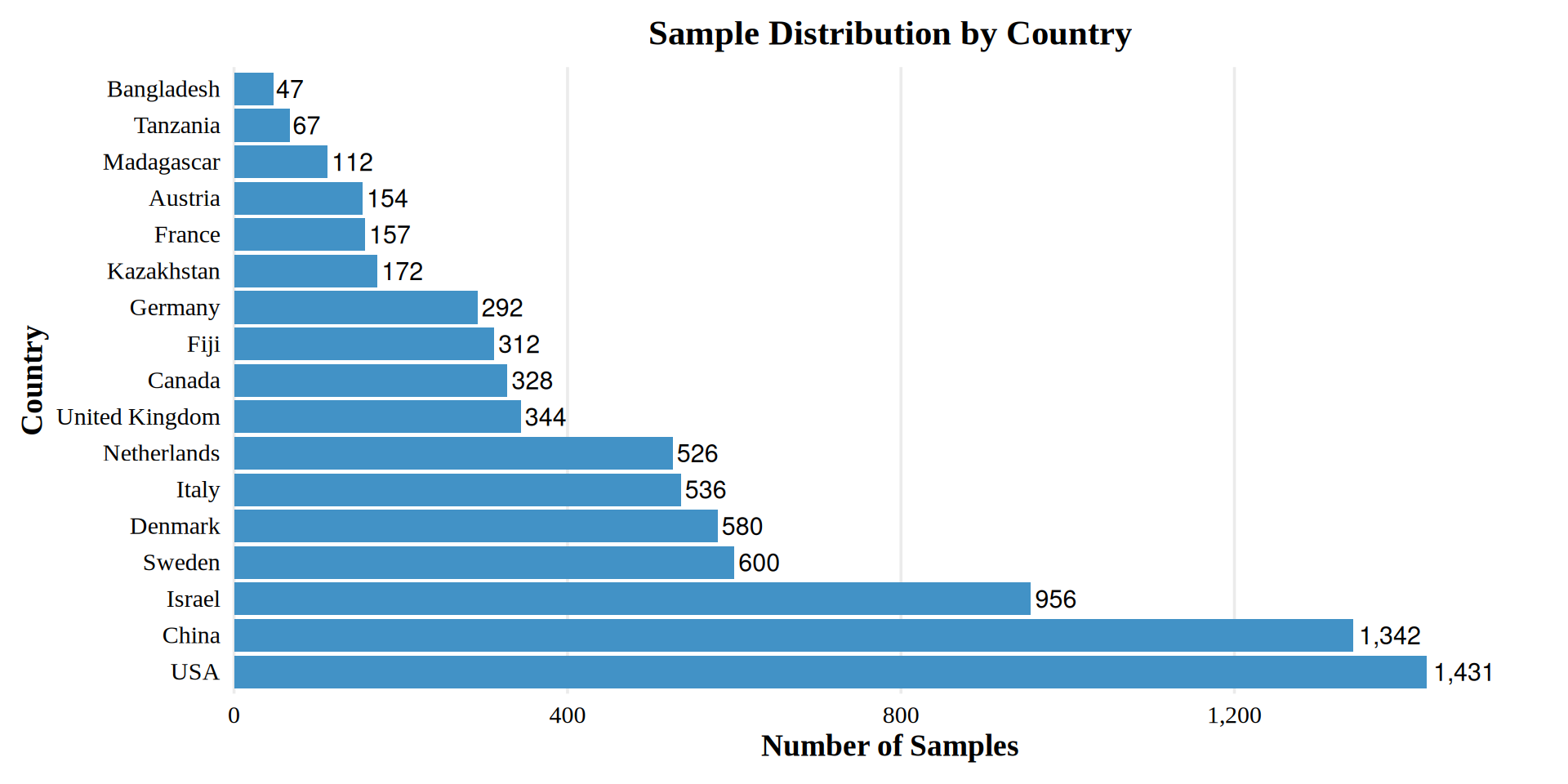
Sample Distribution by Country
Data Subsetting
The data has been carefully subsetted to include samples that meet specific criteria, such as:
Body Site: Limited to stool samples.
Health Status: Focused on healthy individuals not currently using antibiotics.
Age Category: Included only adult subjects.
- SCGs: Selected samples with all 40 Single-Copy Genes (SCGs) recovered .
This subset of the data, now represented as a TreeSummarizedExperiment (TSE) object, is optimized for further analysis and is available for advanced exploration of the antibiotic resistance gene (ARG) load, diversity, and related metadata.
To explore the metadata and access the object, visit this link.
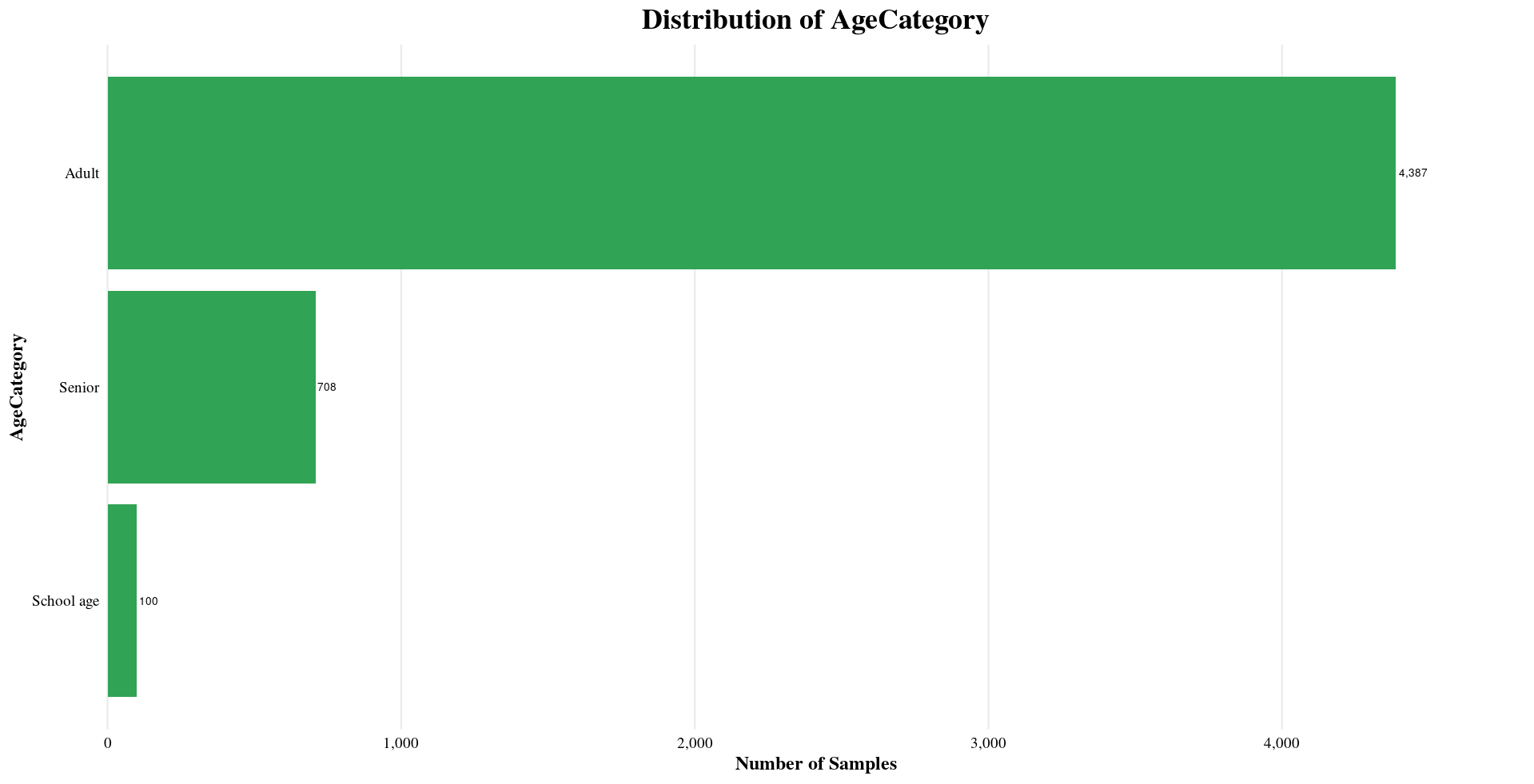
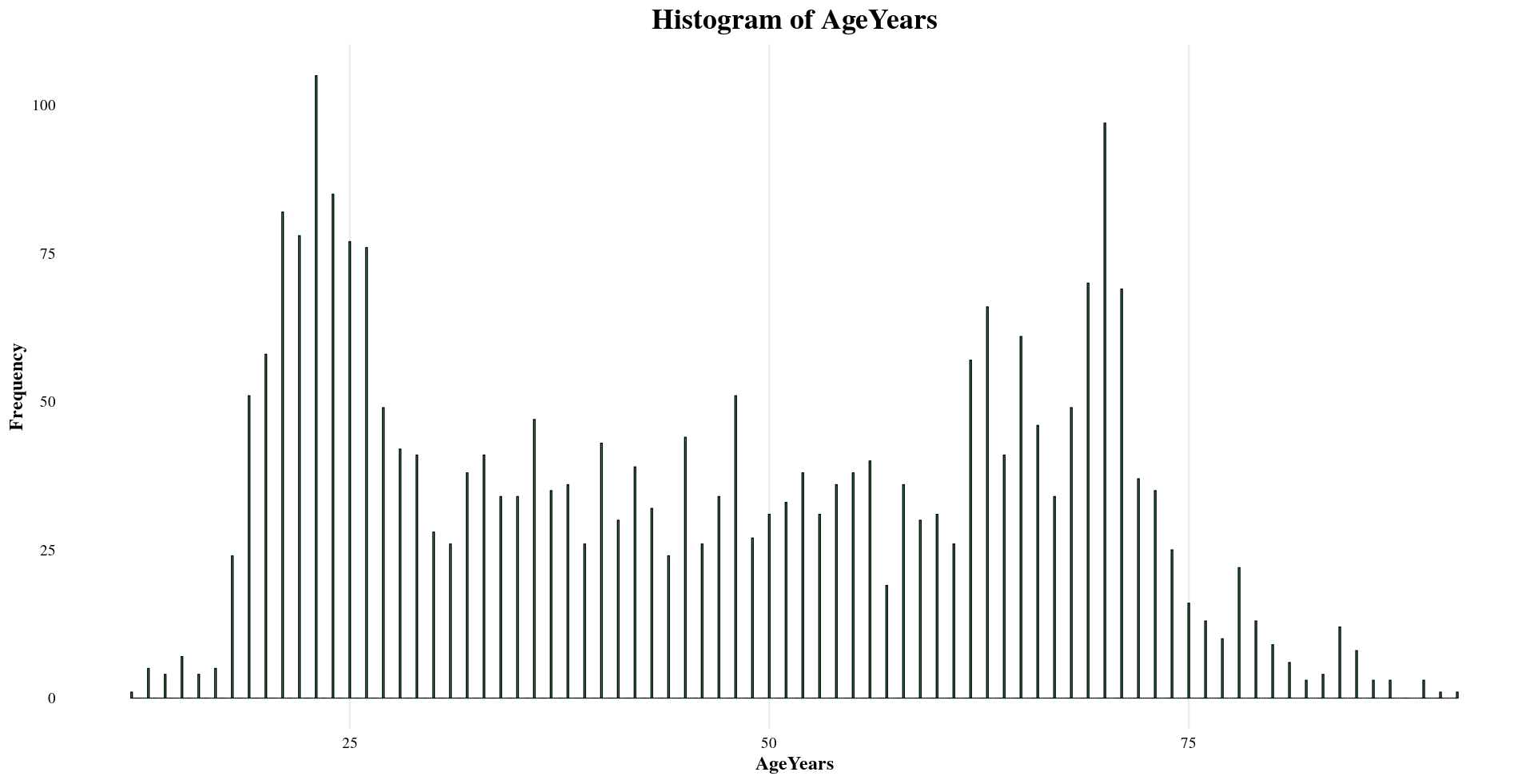
Sample Metadata
The subsetted dataset contains key metadata that provides context to each sample.
Distribution of Sample Metadata
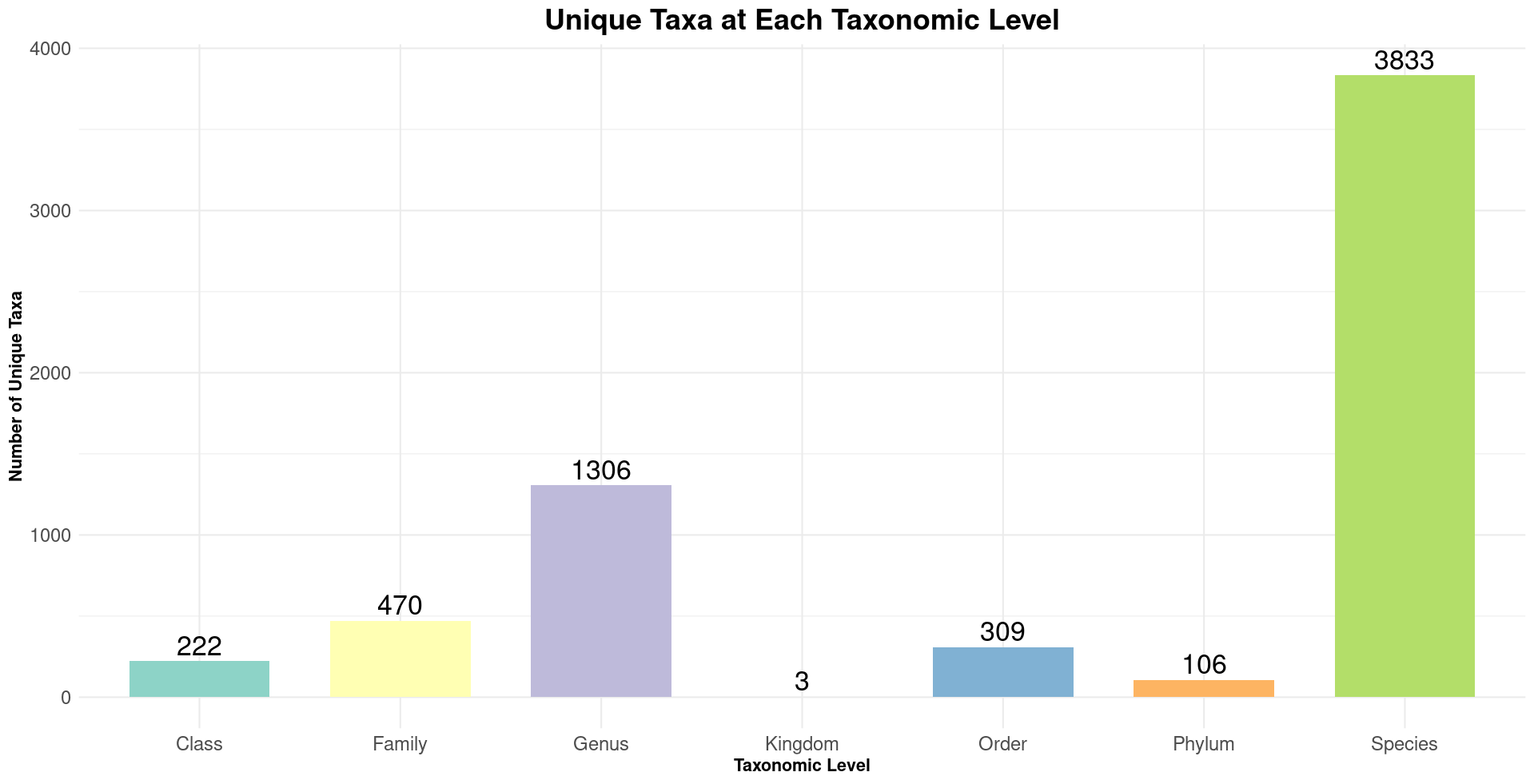
Taxanomic Diversity
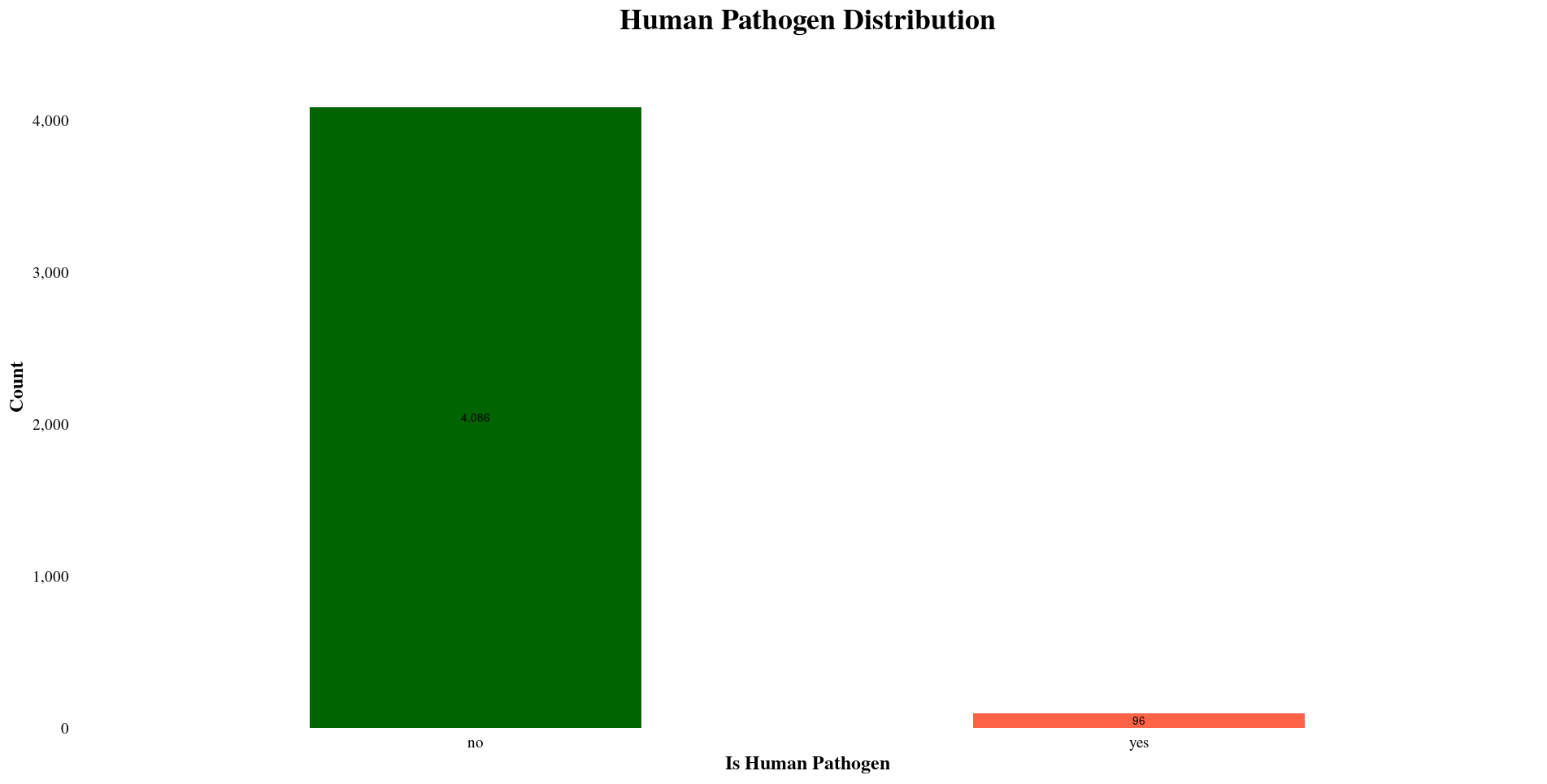
Pathogen Analysis
National Database of Antibiotic Resistant Organisms
National Database of Antibiotic Resistant Organisms (NDARO) is a collaborative, cross-agency, centralized hub for researchers to access AMR data to facilitate real-time surveillance of pathogenic organisms. NDARO is part of the National Action Plan for Combating Antibiotic-Resistant Bacteria developed by the White House in 2015.
NDARO allows:
Identify AMR genes in bacterial genomes with AMRFinder
Identify bacterial genomes with AMR genes in the Isolate Browser
Link to NDARO: https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/