Antimicrobial resistome data analysis with R/Bioconductor
Team Leo Lahti University of Turku, Finland.
Team home page
Data Source
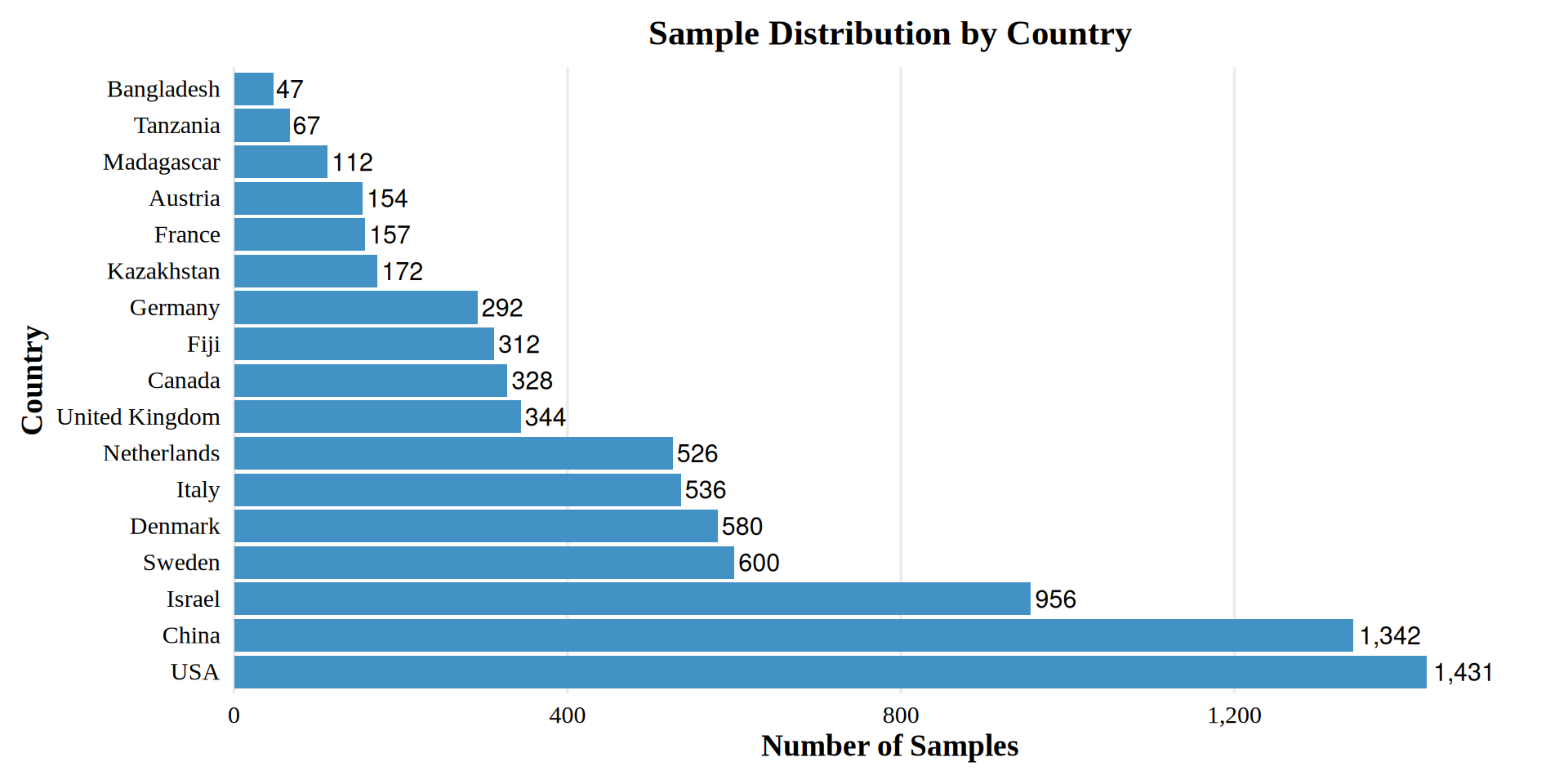
This large-scale study includes a total of 9,251 samples collected from various human body sites and countries.
Raw Illumina sequencing reads from NCBI or EMBL using Kingfisher-download
Metagenomic assemblies retrieved from the paper “Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle”
6104 adult gut metagenome samples from 20 countries after refinning
curatedMetagenomeData R package for retrieving the sample metadata
- Manually curated ARG families from CARD database (n=752)
Sample Distribution by Body Site
The dataset comprises samples from six major body sites, with the distribution as follows:

Sample Distribution by Country
Data overview in R
class: TreeSummarizedExperiment
dim: 4182 5195
metadata(1): Country
assays(1): relabundance
rownames(4182): 362 363 ... 23727 23733
rowData names(13): sgbid sgbname ... assigned_pathogen_name
assigned_pathogen_taxid
colnames(5195): AsnicarF_2017__MV_FEM1_t1Q14
AsnicarF_2017__MV_FEM2_t1Q14 ... ZellerG_2014__CCMD74930188ST-21-0
ZellerG_2014__CCMD79987997ST-21-0
colData names(22): Tier_1.Exclusion_before_analysis
Tier_2.Recover_all_40_SCGs ... antibiotic_exposure_status_descriptive
antibiotic_current_use_binary
reducedDimNames(0):
mainExpName: NULL
altExpNames(2): read assembly
rowLinks: NULL
rowTree: NULL
colLinks: NULL
colTree: NULLPackages used for analysis:
‘mia’
‘miaViz’
‘scater’
‘ComplexHeatmap’
‘pheatmap’
‘dplyr’
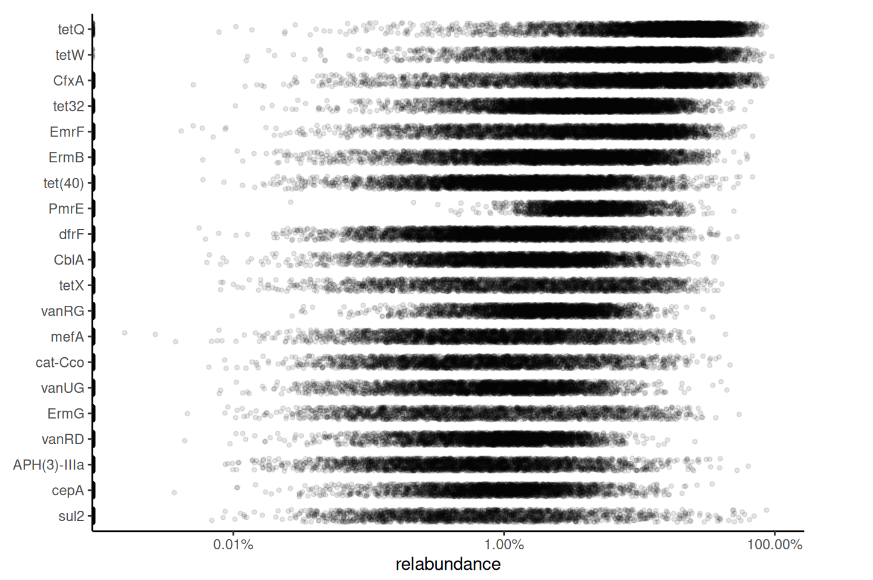
Abundance of resistance genes
Overview of AMR genes
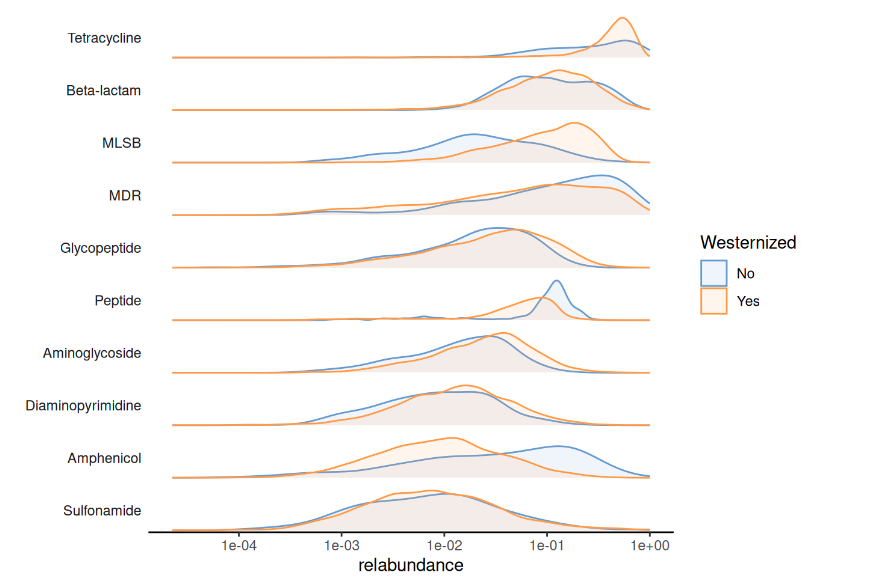
AMR class westernised population
Diversity and Similarity
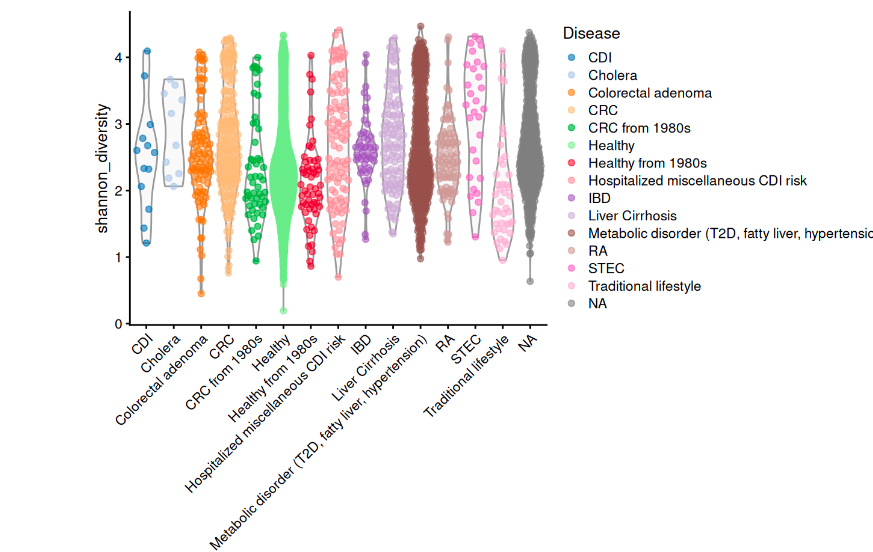
Alpha diversity of AMR genes in different diseases
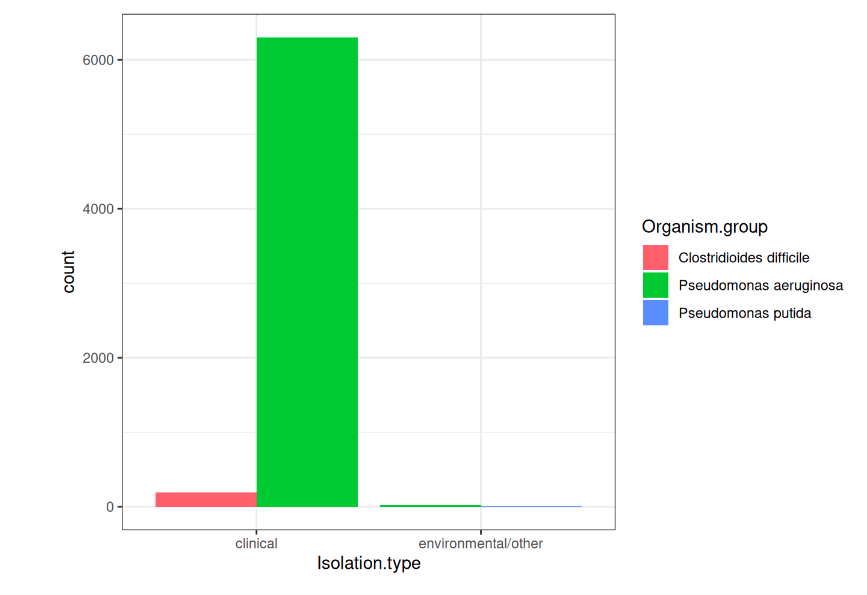
Antibiotic Susceptibility (NDARO)
We analysed the samples from Clostridioides Difficile Infection (CDI) samples and found Pseudomonas aeruginosa was highly prevalent.
Isolation types present in AST data
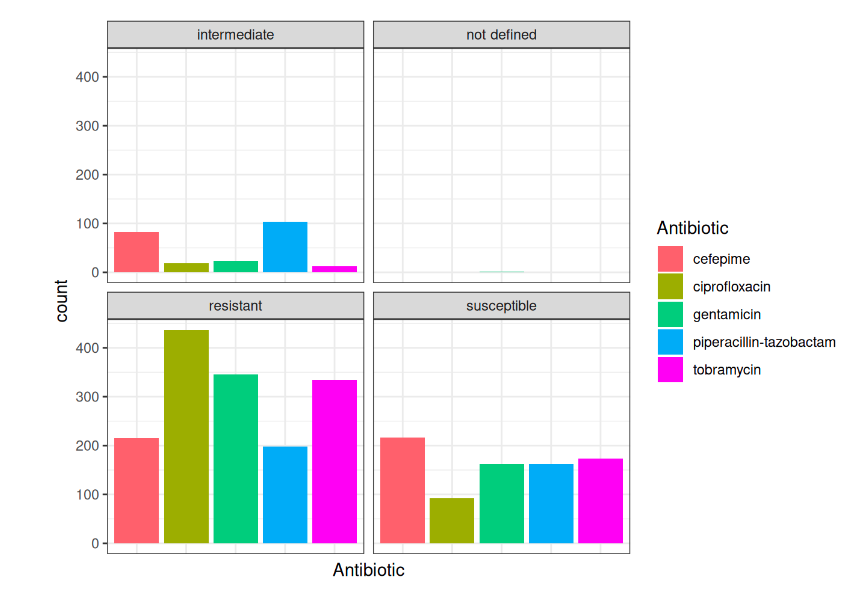
Top-5 antibiotics (with present evidence in AST)
Team Members
Leo Lahti (Team Lead)
Muluh Geraldson (Tech Lead)
Akewak Jeba (Tech Lead)
Nitin Bayal (Writer)
Dattaray Mongad (Writer)
Shivang Bhanushali (Flex)
Mahkameh Salehi (Flex)

