Project goals
This project aims to develop R/Bioconductor tools to analyze AMR data by integrating publicly available human gut metagenomes with ARG profiles (e.g. metagenomics) and epidemiological, transmission data to study AMR gene co-migration.
Approach
Reproducible data science workflow and dissemination material to demonstrate the utilization of the latest Bioconductor multi-assay data integration framework in the context of resistome profiling in large population studies. This covers dedicated data structures and the complementary data analysis and visualization methods. The outcomes are reported via an automated pkgdown website.
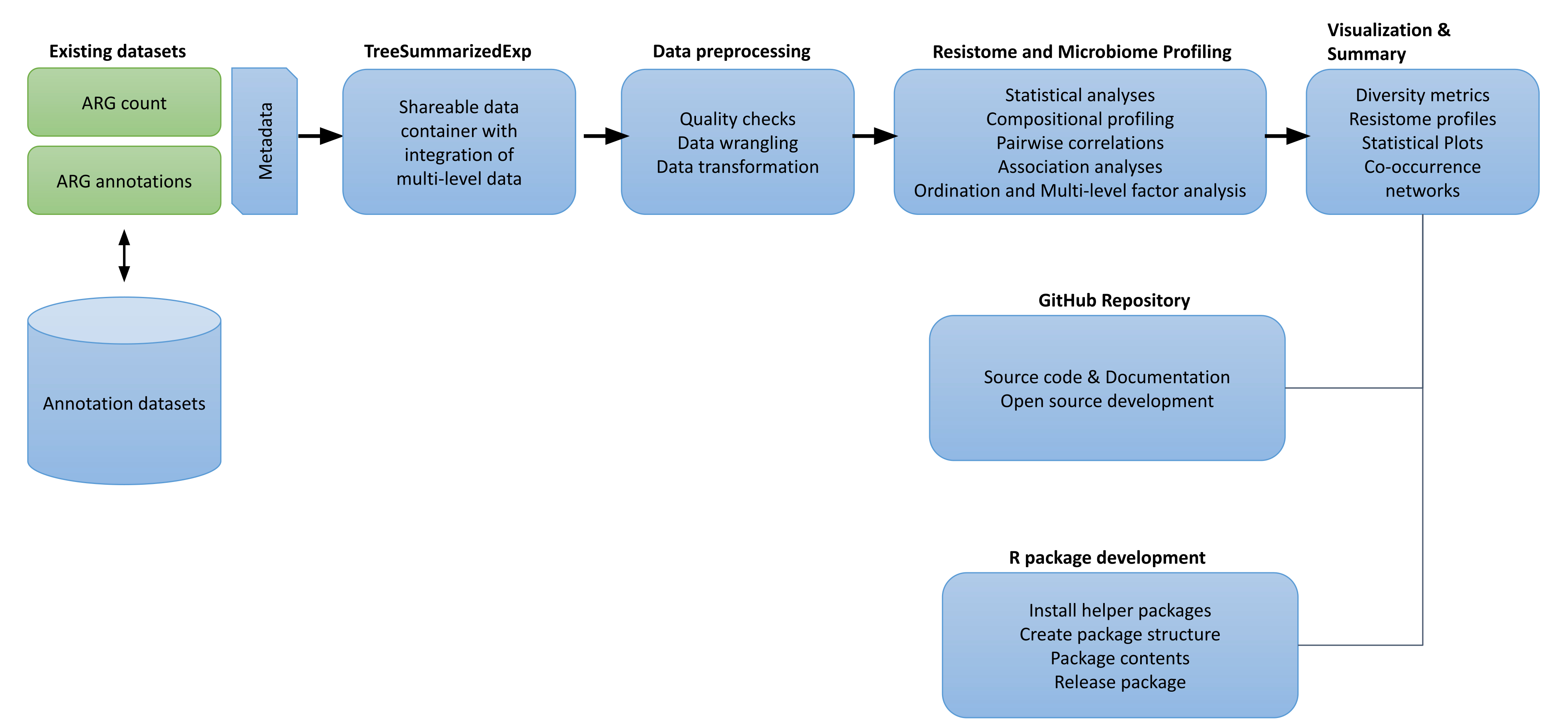
Analysis plan
-
Prepare data
- Prepare the Lee et al. (2023) dataset for analysis in R/Bioc. See data summary.
- Link the dataset to the NDARO database on microbial organisms and resistance genes.
-
Prepare documentation
- Create minimal website for this project.
- Prepare summary of the dataset.
- Create tutorials on resistome data analysis.
- To add documents to the minimal website, see technical guide
-
Analyse the resistome data set
- Provide exploratory summaries of the dataset using R/Bioc methods.
- Replicate some key results from the manuscript to verify the data.
-
Develop methodology
- Implement visualization methods as functions.
-
Finalize workflow
- Polish the data analysis workflow/s & documentation.
Results
The main outcome of this work is the reproducible project site including data science workflows for resistome analysis. This forms the basis for an extendable collection of resistome analysis workflows for reanalysis of data for new discoveries, methods development and educational purposes.
Future work
Future work will standardize and automate some of the key tools, and release them through open source software libraries such as mia in R/Bioconductor.
Team roles
Team Lead / Leo Lahti Conveys scientific objectives to the team, and coordinates work.
Tech Lead / Akewak Jeba & Muluh Geraldson Coordinates software installation and data acquisition, manages version control and the team’s GitHub repository, troubleshoots technical issues with tech support
Writer / Dattaray Mongad & Nitin Bayal Ensures that all work is documented, manages GitHub README and Team Report
Flex / Mahkameh Salehi & Shivang Bhanushali Fills various roles and assumes responsibilities for tasks as needed
NCBI codeathon disclaimer
This software was created as part of an NCBI codeathon(See details) , a hackathon-style event focused on rapid innovation. While we encourage you to explore and adapt this code, please be aware that NCBI does not provide ongoing support for it.
For general questions about NCBI software and tools, please visit: NCBI Contact Page


